Phage therapy: the right virus for the wrong bacteria
Infecting and killing bacteria with phage therapy relies on the use of viruses, so-called bacteriophages, or phages for short. It’s a promising alternative to antibiotics, which are continuing to lose their effectiveness as bacteria develop resistance.
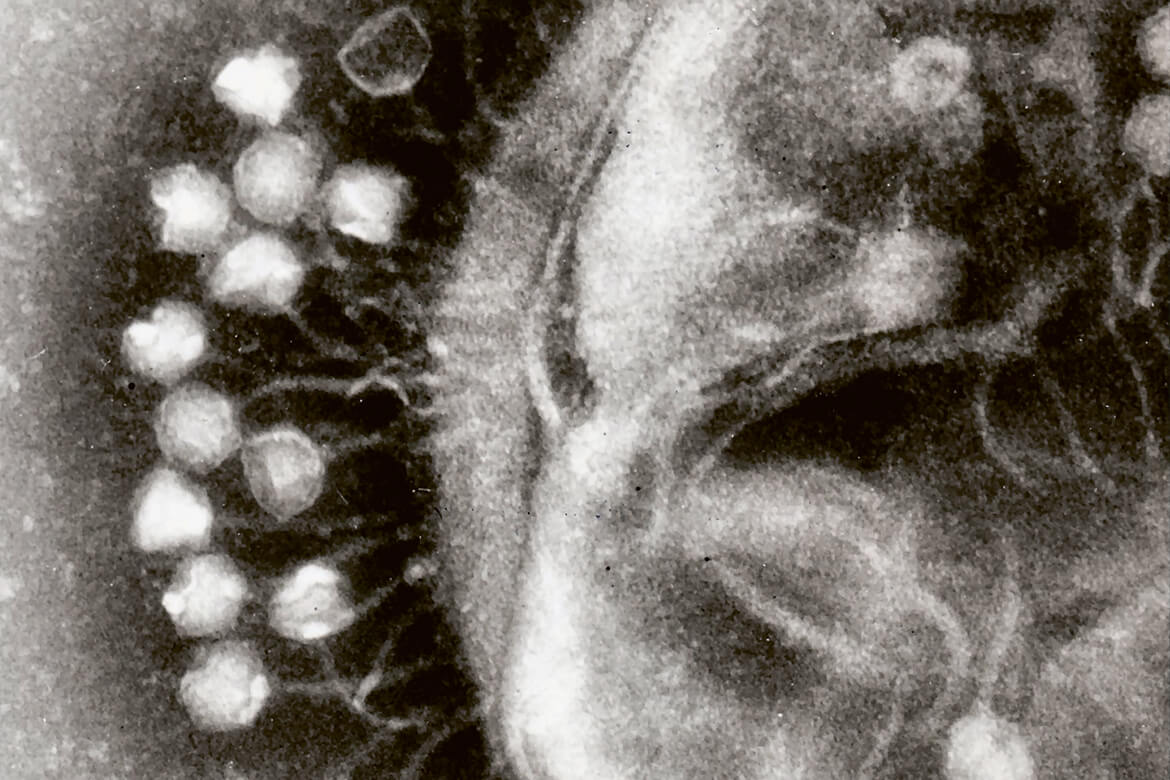
Here, viruses are docking onto the cell wall of a bacterium, and are able to turn it off. | Image: Wikimedia Commons/Dr Graham Beards
Phages are very specific, however, and must be carefully selected if they are to hit their target. Currently, this step involves tedious laboratory testing. To step up the speed of this process, scientists have developed computer models capable of quickly and cost-effectively predicting phage-bacteria interactions based on the analysis of their genomes. The study was carried out by the Swiss Institute of Bioinformatics (SIB), the School of Management and Engineering Vaud, the University of Lausanne and the University Hospital of Bern.
The predictive models employ supervised automatic learning techniques, a statistical approach that allows an algorithm to learn from data. “We collected information on more than a thousand interactions between phages and bacteria for which there are complete genome sequences”, says Carlos Peña of the SIB, who led the study. From these genomes, the scientists then extracted indicators for the specific characteristics of phages and bacteria, such as the structure of certain proteins and whether they bind to each other. This was the data used to train the algorithm. It can correctly predict about 90 percent of interactions. “Of course, there’s no way around having to test the predictions in the laboratory, but it’s now much more targeted and saves us a considerable amount of time”.
D.M.C. Leite et al.: Computational prediction of inter-species relationships through omics data analysis and machine learning. BMC Bioinformatics (2018)